Copyright © 2007-2010 BiochemLabSolutions.com
Wouldn't it be great to be able to draw any molecule you can imagine and dock it flexibly to a protein of your choice from the PDB
database?
Can be done.
database?
Can be done.


Step 1: draw a molecule, convert it to 3D and save as *.MOL
To draw a chemical structure, you can use ChemSketch program.
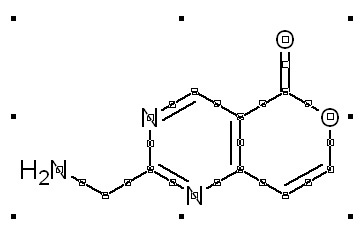
Manual drawing of a molecule from scratch in ChemSketch
3D conversion in ChemSketch

Step 2: convert *.MOL to *.PDB
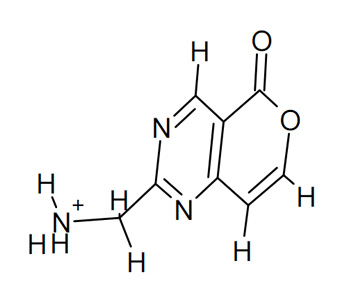
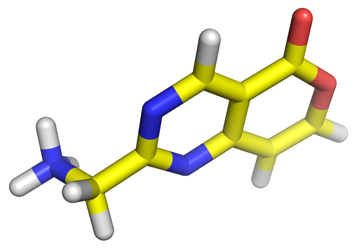

Step 3: prepare PDBQT of the ligand and the protein
Use MGLTools or our conversion utility to prepare PDBQT files from the PDB files of your above ligand and protein target of your choice from the PDB data bank.

Step 4: dock the small molecule to your protein
For this step you can use VcPpt. Drag and Drop the protein and ligand PDBQT files and click RUN.
The protein of choice this time will be HIV reverse transcriptase, PDB ID 3MEC.
The protein of choice this time will be HIV reverse transcriptase, PDB ID 3MEC.

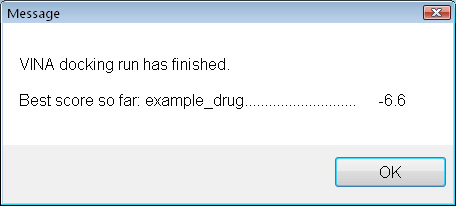
Once the docking is complete, it seems that this small molecule does not bind very strongly. A score of -9 to -12 would be expacted for a well-binding ligand. How about adding a few carbon atoms ?

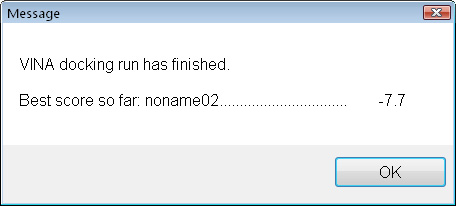
Still not great, but definitely better. Let's look where it bound and how.
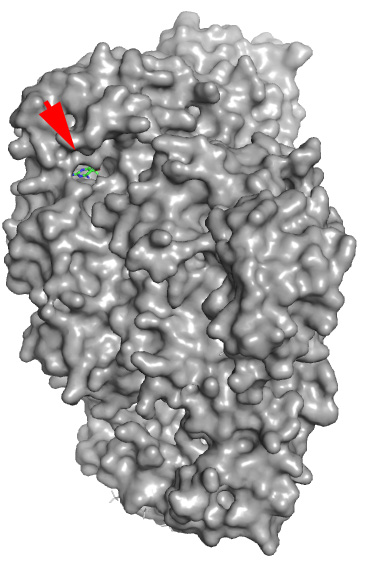
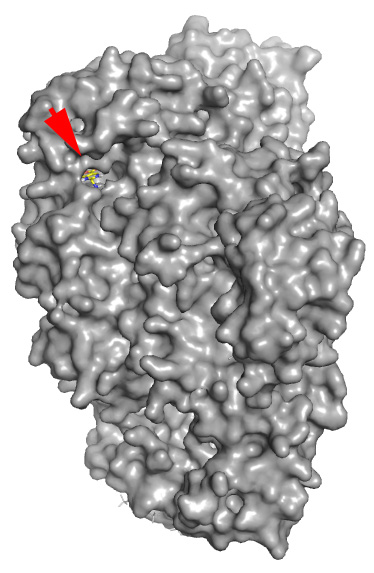
Real structure with co-crystallized Docking prediction with the example
ligand (shown for reference) drug just drawn in ChemSketch - note that
the molecule was docked in the same pocket
without any prior knowledge
ligand (shown for reference) drug just drawn in ChemSketch - note that
the molecule was docked in the same pocket
without any prior knowledge

Step 5: look at the results
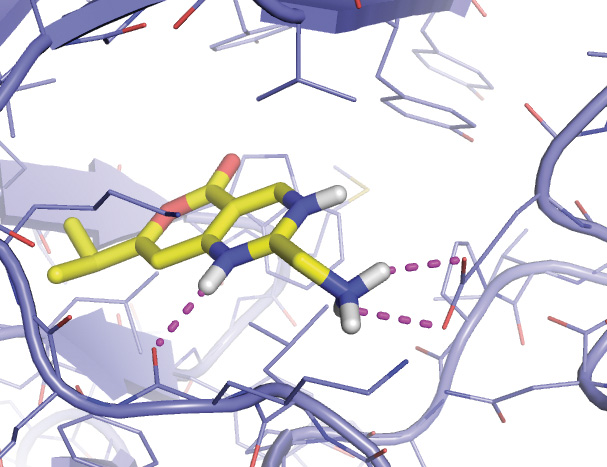
The red arrow shows where real and fantasy small molecules are bound.
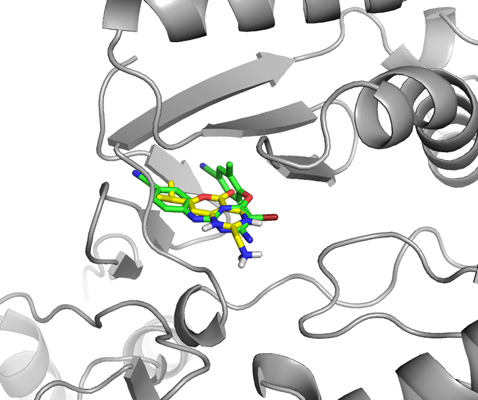
Close up view of both ligands, the one from the actual crystal structure and the one drawn in ChemSketch from scratch.
Final close up view of the ligand drawn in ChemSketch docked to HIV reverse transcriptase.