Copyright © 2007-2010 BiochemLabSolutions.com
VcPpt is an independently developed extension for Vina, a program for flexible ligand docking under Windows OS
Carry out high-throughput screening of compound libraries with Vina and cluster resulting PDB files of ligands by binding energies and by binding positions. Results processing functions do not require Vina installed. Screening scale is unlimited.
Carry out high-throughput screening of compound libraries with Vina and cluster resulting PDB files of ligands by binding energies and by binding positions. Results processing functions do not require Vina installed. Screening scale is unlimited.

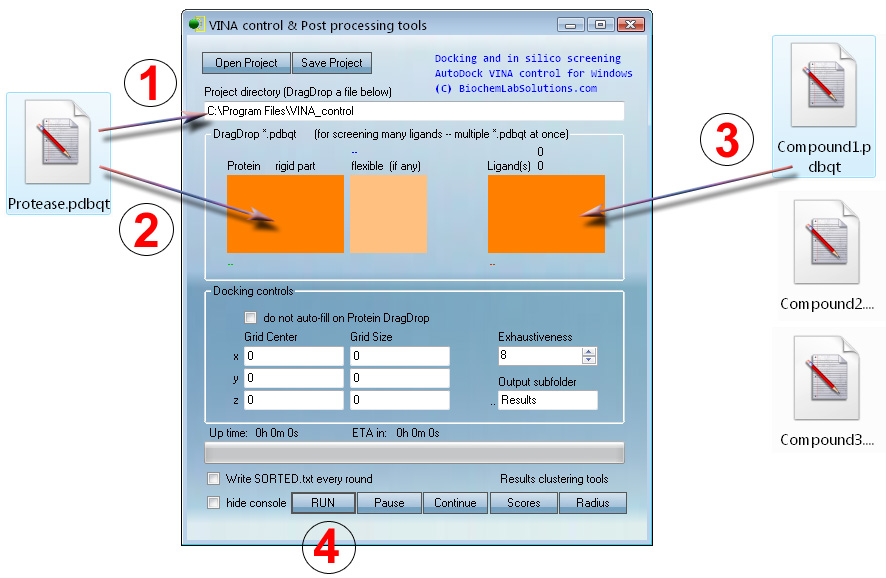
Note that grid parameters are automatically computed at step 1 to cover the entire Protease.pdbqt. Optionally (for advanced users) - to focus fitting on a specific protein pocket - enter the x, y, z manually.
At step 3, any number of ligands (100,000 ? - sure) can be drag-dropped for automatic screening.
"Grid box" is a selection of 3D space around the protein.
Ligands are being moved and docked only within constraints of this 3D box.
Box center = "grid center", box size = "grid size".
At step 3, any number of ligands (100,000 ? - sure) can be drag-dropped for automatic screening.
"Grid box" is a selection of 3D space around the protein.
Ligands are being moved and docked only within constraints of this 3D box.
Box center = "grid center", box size = "grid size".

Docking or screening with VcPpt can be set up in four mouse moves:
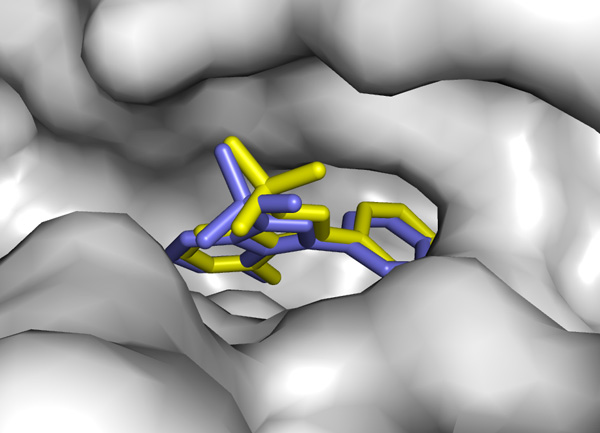

How good can docking predictions be? See this calculation vs a real Xray structure:
"not bad at all" result thanks to the algorithm in vina.exe
For more information on VINA, pdbqt and on how to convert PDB, mol, SMILES
or other files to pdbqt, please visit MGL tools web site and read our formats page.
Search keywords
drug discovery, drug targeting, medicine, biomedicine, pharmacology software,
drug development, small molecule inhibitors
For more information on VINA, pdbqt and on how to convert PDB, mol, SMILES
or other files to pdbqt, please visit MGL tools web site and read our formats page.
Search keywords
drug discovery, drug targeting, medicine, biomedicine, pharmacology software,
drug development, small molecule inhibitors

What you need to actually screen first hundreds of small molecules against
your PDB, on your Windows computer, today:
minimum software.........................VcPpt Vina.exe MGLTools
optional software...........................EasyConvert OpenBabel
+ small molecule data sets in any format from here here or here
Usual sequence of actions:
> Download small molecules as a single SDF file
> Convert SDF to many PDB files in EasyConvert
> Convert PDB files to PDBqt files in EasyConvert
> Convert your protein PDB to PDBqt in EasyConvert
> Dock in VcPpt
> Sort docking results in VcPpt using sorting and clustering tools
> View docking results in PyMol or in UCSF Chimera
your PDB, on your Windows computer, today:
minimum software.........................VcPpt Vina.exe MGLTools
optional software...........................EasyConvert OpenBabel
+ small molecule data sets in any format from here here or here
Usual sequence of actions:
> Download small molecules as a single SDF file
> Convert SDF to many PDB files in EasyConvert
> Convert PDB files to PDBqt files in EasyConvert
> Convert your protein PDB to PDBqt in EasyConvert
> Dock in VcPpt
> Sort docking results in VcPpt using sorting and clustering tools
> View docking results in PyMol or in UCSF Chimera