Copyright © 2007-2010 BiochemLabSolutions.com
Vina and VcPpt require protein and small molecule ligands to be in .PDBQT format. Traditional file formats SMILES, SDF, PDB and MOL can be converted to work with VcPpt.

A good source of small molecule libraries UCSF ZINC database. Millions of its structures can be converted to .pdbqt format and screened with VcPpt. This database contains 3D-optimized structures in SDF format and contains, among others, FDA approved compounds, Sigma and Acros compounds and compounds from major screening libraries.

Small molecule database

Converting ligand file formats
SDF to PDB (sdf2pdb)
MOL to PDB (mol2pdb)
PDB to MOL (pdb2mol)
These conversions can be done via command line or GUI in Open Babel.
Note about SMILES and SDF formats: SMILES is 2D format and SDF may contain either 2D or 3D structures. In case SDF files come in 3D, as in ZINC database, Open Babel will handle them well and convert to PDB correctly. If SDF 2D or SMILES strings are used, a 2Dto3D conversion must be run.
MOL to PDB (mol2pdb)
PDB to MOL (pdb2mol)
These conversions can be done via command line or GUI in Open Babel.
Note about SMILES and SDF formats: SMILES is 2D format and SDF may contain either 2D or 3D structures. In case SDF files come in 3D, as in ZINC database, Open Babel will handle them well and convert to PDB correctly. If SDF 2D or SMILES strings are used, a 2Dto3D conversion must be run.


"Easy Convert"
Instant Drag and Drop batch conversion
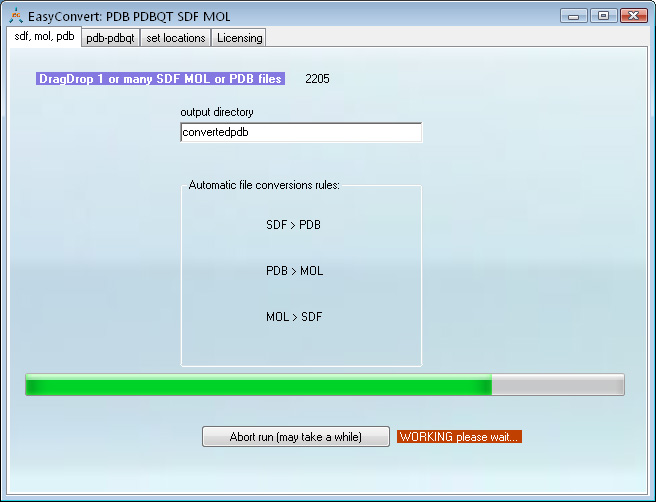
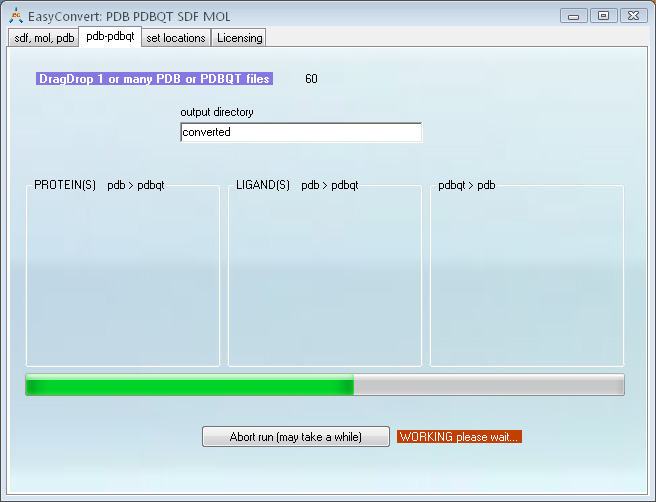
This program uses Open Babel, MGLTools (which you need to download separately) as well as its own built-in conversion algorithms.
Conversion between sdf, pdb, pdbqt and mol is done automatically upon DragDrop of the input files. The program will convert as many files as were DragDropped at once
(10000 - sure).
Introduced in V 1.5:
3D optimization option to convert any flat 2D sdf, pdb or mol structures to 3D
-- requires OpenBabel 2.2.3 or 2.3.0 or better
Introduced in V 1.6 (August 2011)
Protein PDBQT generation works correctly with problematic PDBs
Conversion between sdf, pdb, pdbqt and mol is done automatically upon DragDrop of the input files. The program will convert as many files as were DragDropped at once
(10000 - sure).
Introduced in V 1.5:
3D optimization option to convert any flat 2D sdf, pdb or mol structures to 3D
-- requires OpenBabel 2.2.3 or 2.3.0 or better
Introduced in V 1.6 (August 2011)
Protein PDBQT generation works correctly with problematic PDBs
Screen shot
Screen shot