


Basic docking or screening with VcPpt can be set up in just four mouse moves:
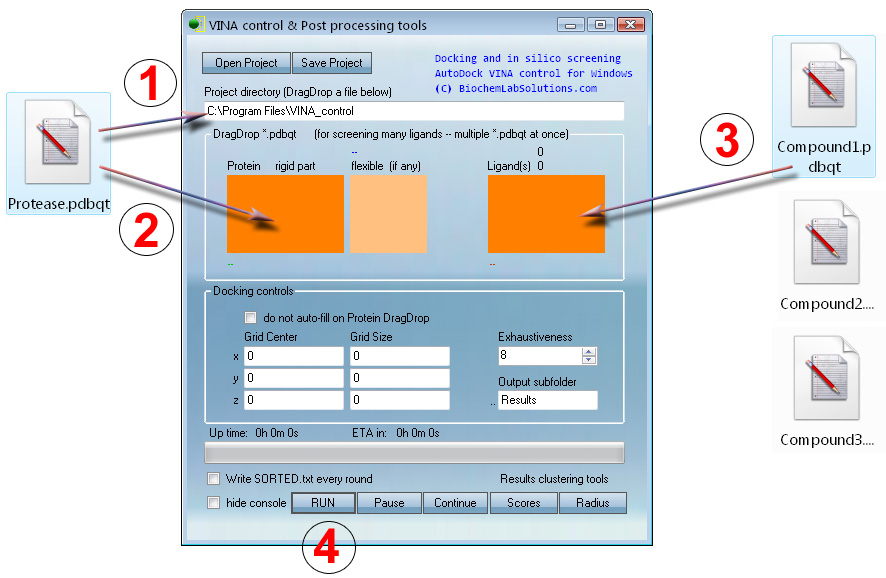
Note that grid parameters are automatically computed at
step 1 to cover the
entire Protease.pdbqt.
Optionally (for advanced users) - to focus fitting on a
specific protein pocket, enter the x, y, z manually.
At step 3, any number of ligands (100,000 ? - sure) can be
drag-dropped for automatic screening.
"Grid box" is a selection of 3D space around the protein.
Ligands are being moved and docked only within constraints
of this 3D box.
Box center = "grid center", box size = "grid size".
Results sorting tools
Docking results from 20 small molecule ligands (as on this
picture) can be managed very easily.
As count grows to 1000 or <much> more, sorting and clustering of the docked
solutions becomes
the primary task.
VcPpt contains all the tools for very rapid processing of the docked
results to group them by energy,
binding pockets, or both. Vina.exe is not required for these tasks and
need not be installed.
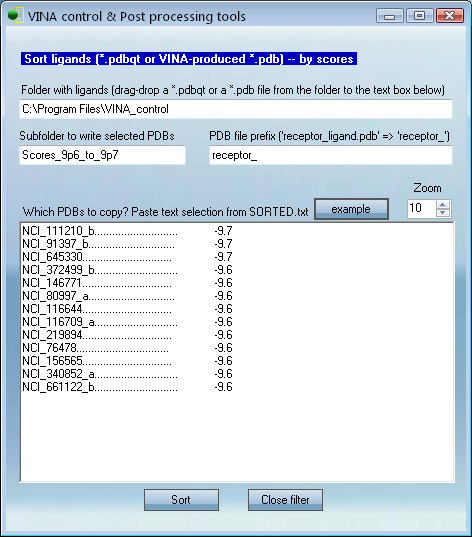
This panel is useful for finding the best binders from
in silico high
throughput screening. An in silico screen may result in 10,000+ pdb
files, most of which will have poor scores and need to be
omitted from further analysis. Here one can find top binders and copy
them to a new subfolder through a simple procedure.
1. Provide folder with
screening results in place of "C:\" .
2. Give subfolder name for placing filtered PDB
results in place of "Scores..."
3. PDB files from docking/screening run have file
names in the form
"receptor_ligand1.pdb",
"receptor_ligand2.pdb",
"receptor_ligand3.pdb",
.....
"receptor" is the name of the
protein PDBQT file used in the screening
"ligand1", "ligand2".... are the
names of the small molecules PDBQT
At this step the program will
need to know the protein name used.
For example, If the screening was
done with "kinase.pdbqt", "kinase_" should be typed instead of "receptor_".
4. Final large text box contains the filtering rules.
Ligands listed there provide the guide for
finding and copying PDBs to the
filtered subfolder.
The whole-ligand list is automatically
generated during the docking run and is called "SORTED.txt".
One should open SORTED.txt in a text
editor and copy any number of entries (for example, top 100)
to the filtering rules window. When
copying, avoid empty lines at the list's end. Each line should have
a ligand.
5. The "Sort" button will run the sorting job.
If "SORTED.txt" is not available (for example, you
combined separate VcPpt screening results into
one folder, or have Vina docking results made not* with
VcPpt), it is easy to make new SORTED.txt.
1. In VcPpt, click "Radius".
2. Drag-Drop one of the PDBs to the orange pad (this will
point VcPpt to the right working directory with Vina outputs)
3. In "Make SORTED.txt for any folder with VINA
PDBs", PDBQT protein name with "_" goes instead of "receptor_"
(as in step 3 above)
4. "Make" will prepare new "SORTED.txt" in the docked
ligands directory.
*If PDB docking files were made not with VcPpt, these PDB
files must be renamed to contain a common prefix.
For example if one's Vina output files at hand are:
"drug.pdb", "adenine.pdb", "19000.pdb",
one renames them to "new_drug.pdb", "new_adenine.pdb",
"new_19000.pdb" and
replaces the word "receptor_" with "new_" in the text box
of "Make SORTED.txt for any folder with VINA PDBs".
The "Make" button will produce "SORTED.txt".
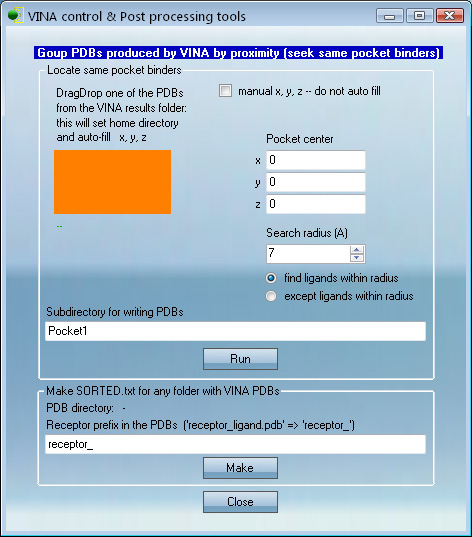
This panel works similar but finds ligands within some
distance from a certain point in 3D space.
For example, if one has a ligand and needs to find which
other ligands were docked nearby.
1. Drag-Drop this ligand's PDB to the orange pad; x, y and
z will be filled pointing at the
Drag-Dropped ligand's center.
PDBs to be searched must be present in
the same folder with this ligand.
2. The "RUN" button will find PDBs whose top scoring
solution is within 7A (distance is adjustable)
from the x, y, z. These PDBs will be
copied to the subfolder "Pocket1".
- x, y, z can be entered manually
- x, y, z can also be computed by copying a few amino
acids from the pocket of interest in target protein
to a new PDB file and Drag-Dropping this file to the
orange pad to set x, y, and z of the pocket.
"Manual x, y, z -- do not auto fill" should then be
checked to fix the coordinates.
Then Steps 1 and 2 above are done as usual.
All sorting is done based on the top docked solution in
the PDB files.
By combining the sorting by scores and sorting by binding
position it takes minutes to find the best binders to
any given pocket among thousands of docking results.
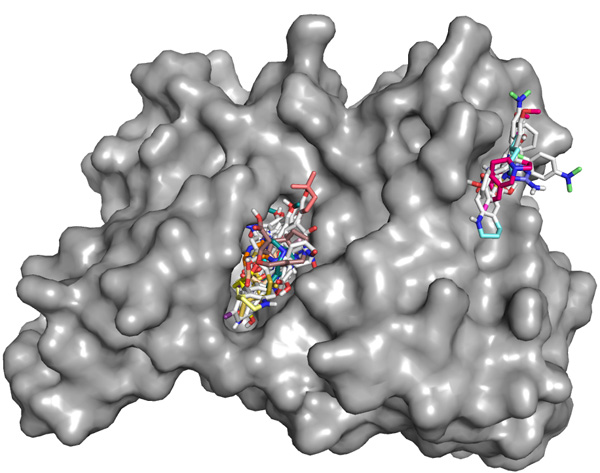
How good can docking predictions be? See this calculation
vs a real Xray structure:
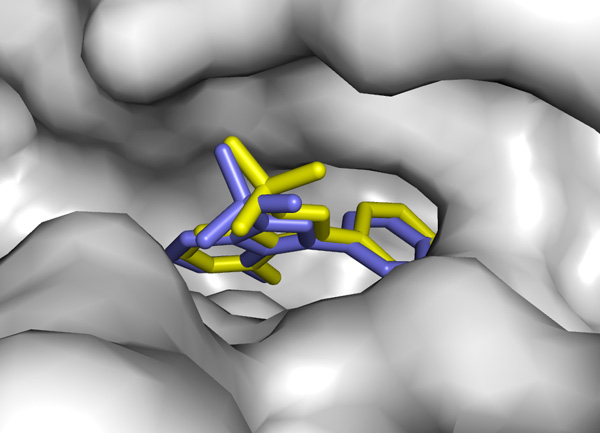
"not so bad at all" result thanks to the algorithm in
vina.exe
For more information on VINA, pdbqt and on how to convert
PDB, mol, SMILES
or other files to pdbqt, please visit
MGL tools web site.
A good source of small molecule libraries UCSF
ZINC database. Millions of its
structures can easily
be converted to .pdbqt format and screened with VcPpt.
Interested in our pdb > pdbqt; SDF > pdbqt batch conversion tools or in
drug-likeliness
ligand sorting tools?
Inquire by
email.
This program is an independent extension for VINA. This
site is not affiliated with Vina developers.
Download
The software is provided with limited functionality,
namely
(1) docking is limited to 3 ligands at a time
(2) running time is limited to 20 minutes per use
To activate unlimited number of ligands and running time
users should use built-in registration tool.
Other downloads one may need for the program to work:
(1)
.NET 3.5
(2) Sorting and clustering
functions will work out of the box [1].
However, Vina must be obtained if you wish to carry out docking and in
silico screening.
Supported OS:
Windows XP, Vista 32/64 bit, Windows Server 2003 and 2008, Windows 7.
.NET 3.5 must be installed for the program to run. Otherwise, an error code will
appear.
A note about UAC of Windows: you may see an error
message when starting
the program if your UAC is turned on because Administrative priviliges are
required to run.
Solution 1:
find C:\Program Files\VINA_control\VINA_control.exe,
right-click the exe file
find Compatibility->Privilege Level
check "Run this program as Administrator", done.
Windows 7 and Vista users: please note the (resolved) DragDrop issue
in certain usage scenarios.
Solution 2 (quick)
Right-click program icon, select "Run as administrator"
Please don't forget to read the FAQ section about usage
scenarios
MacOS and
Linux users can run this program at native speed via
free
VirtualBox by Sun
Microsystems.
[1] To block automatic prompt for "vina.exe"
download in case you do not have or can't have VINA:
- create an empty text file in the VcPpt install directory
- name it "vina.exe"
Contact:
eln biochemlabsolutions.com
biochemlabsolutions.com
Copyright © 2009-2010
BiochemLabSolutions.com