
Index

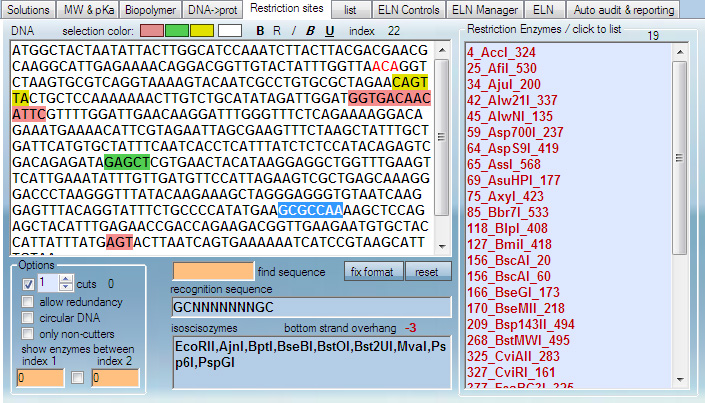
This module finds restriction sites in DNA. The operation of the module is straightforward, except that to find enzymes that cut DNA any number of times, "0 cuts" should be selected.
This module works interactively, reading mouse position in real time to indicate restriction sites.
THE USE OF RESTRICTION SITES FINDER
DNA sequence is pasted in the main window. Enzymes that cut a certain number of times are shown in the left box. Hovering mouse over any enzyme highlights its recognition site in DNA sequence and shows the enzyme summary (isoscisozymes, produced overhang etc).
If recognition site in DNA is not highlighted, click the DNA sequence once, then hover the mose over enzymes.
It is possible to find enzymes that cut only between specific indexes in the DNA sequence.
Clicking restriction enzymes from the list box places them to the list menu for further use.
SAVING THE WORK
Formatted results can be saved to *.dca files for later use.