
Index

This module calculates Mw, extinction coefficients and concentrations of DNA, RNA and proteins from their sequence (and UV absorbance at 260 nM (RNA/DNA) or 280 nM (proteins)). For proteins, isoelectric points are also calculated.
RNA/DNA EXTINCTION COEFFICIENTS
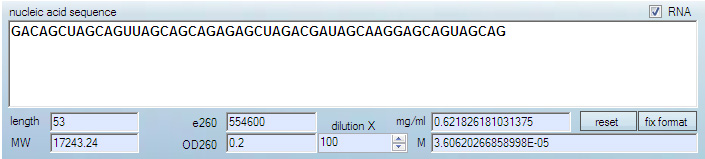
For DNA and RNA, nearest neighbor approach is used. Extinction coefficients that result are rather precise.
Note that secondary structure may interfere with extinction coefficients of nucleic acids, often making them absorb ~20-25% less than unstructured oligonucleotides.
PROTEIN EXTINCTION COEFFICIENTS
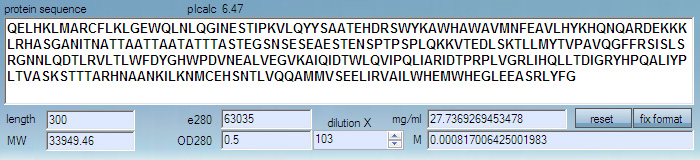
For proteins, W + Y + C absorbance is used, giving very good predictions, especially when protein absorbance is measured in 6.0 M guanidinium hydrochloride, 0.02 M phosphate buffer, pH 6.5.
In practice, absorbance of many proteins is accurate even if measured in pure water.