
Chemical similarity scoring and search tutorial
MassiveSAR provides a simple way for finding structurally similar or structurally related molecules in chemical data sets. For example, when conducting high-throughput screens, it may be important to find molecules within 80% of structural similarity to a certain good hit. This could be used to find out which molecules look similar and work well and which molecules look similar and do not work - to establish structure-activity relationships, SAR.
MassiveSAR provides a simple way for finding structurally similar or structurally related molecules in chemical data sets. For example, when conducting high-throughput screens, it may be important to find molecules within 80% of structural similarity to a certain good hit. This could be used to find out which molecules look similar and work well and which molecules look similar and do not work - to establish structure-activity relationships, SAR.

Step 1. Download SDF database from UCSF ZINC site
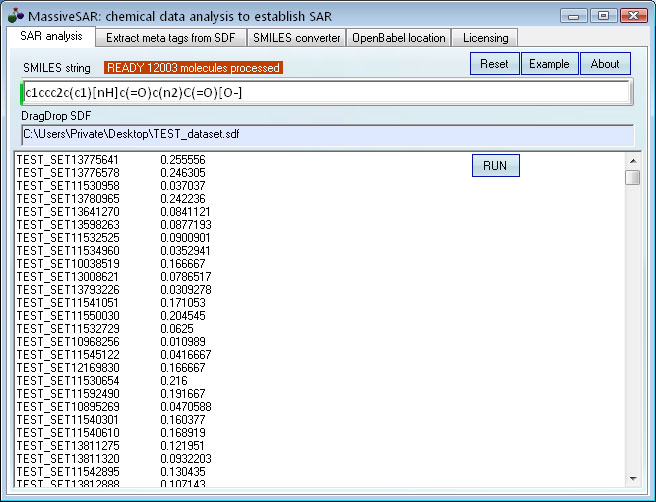
[O-]C(=O)C1=Nc2ccccc2NC1=O
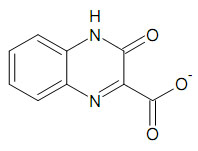

Example: find compounds similar to this key structure among ~12,000 hits in Acros SDF database.
SMILES notation:
Any SDF dataset can be used for SAR analysis. Of course, most interesting are datasets produced by actual "wet" high-throughput screening. For this excersize, however, we will use Acros data set.

Step 2. Run similarity scoring using MassiveSAR and SMILES notation of the key compound
At this step, query compound provided as SMILES text will be compared to every molecule in the SDF file and scored for chemical similarity.
Set up scoring run as follows. Running time should be < 1 min for ~ 10,000 molecules on a modern CPU.
Set up scoring run as follows. Running time should be < 1 min for ~ 10,000 molecules on a modern CPU.

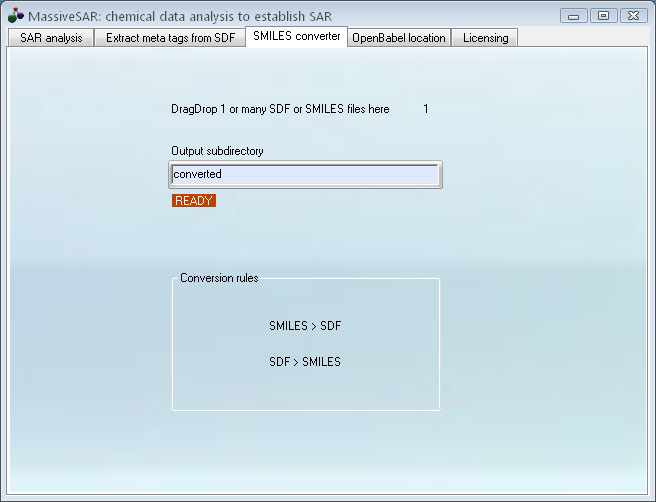
Step 3. Convert the SDF data to SMILES for pasting to Excel
DragDrop SDF file onto the program.
Copyright © 2007-2010 BiochemLabSolutions.com

Resulting SMILES file in "./converted" subfolder:

Step 4. Open resulting smi file in Excel
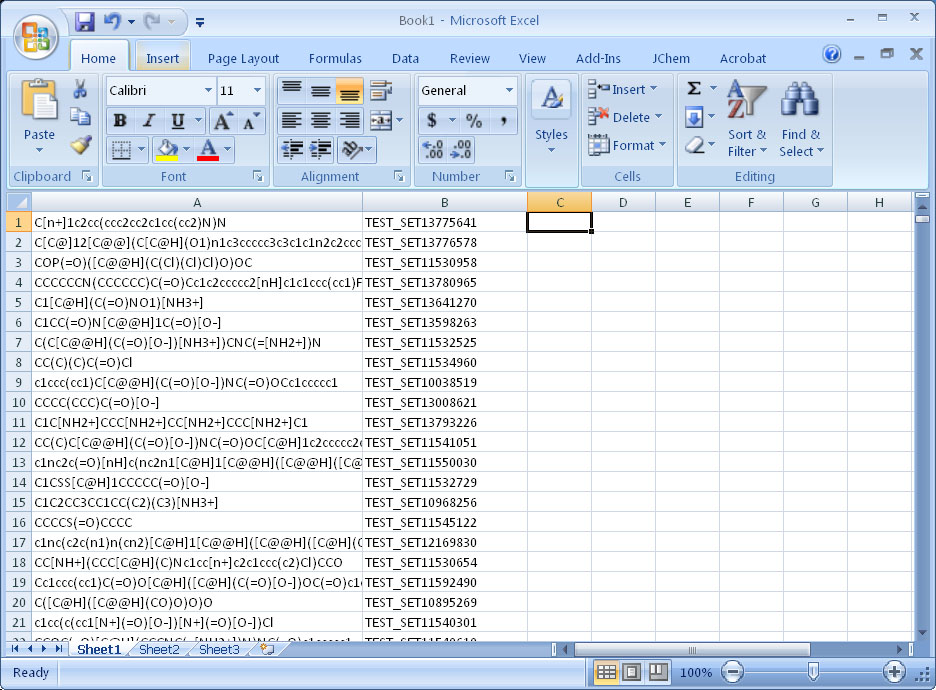

Step 5. Add scores produced in Step 2; sort the data
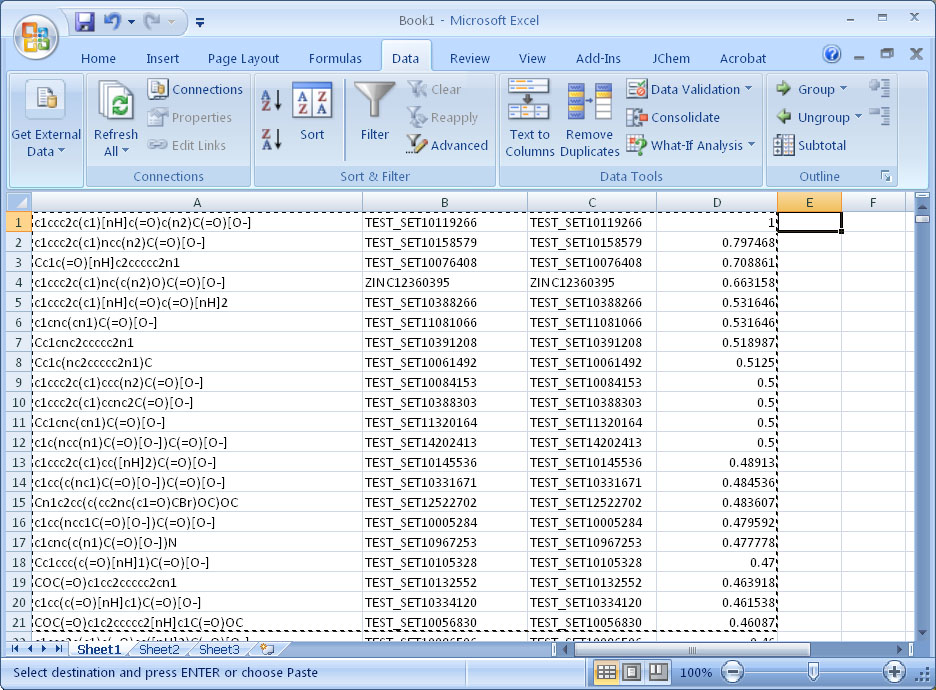
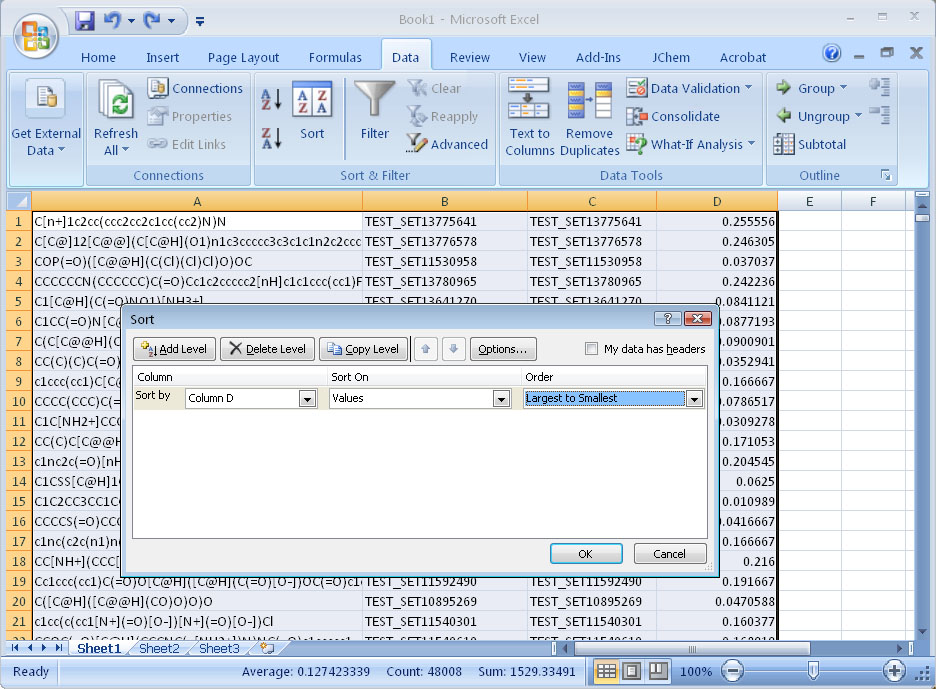

Step 6. Examine the results
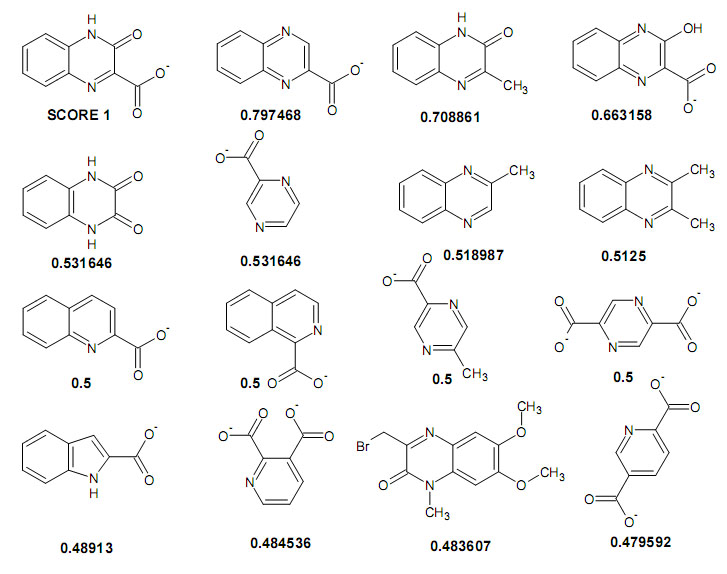
To view structures, one could use JChem or ChemSketch (use Shift+Ctrl+U to paste SMILES strings directly to ChemSketch). First 16 most query-like molecules (out of total of 12,004 molecules analyzed) are shown:












