
SEQMOL index

DNA sequencing tool
Download
This tool allows to semi-automatically assemble DNA sequencing results into a complete sequence. The utility particularly speeds up cumbersome sequence assemblies, for example, when several forward and reverse primers were used.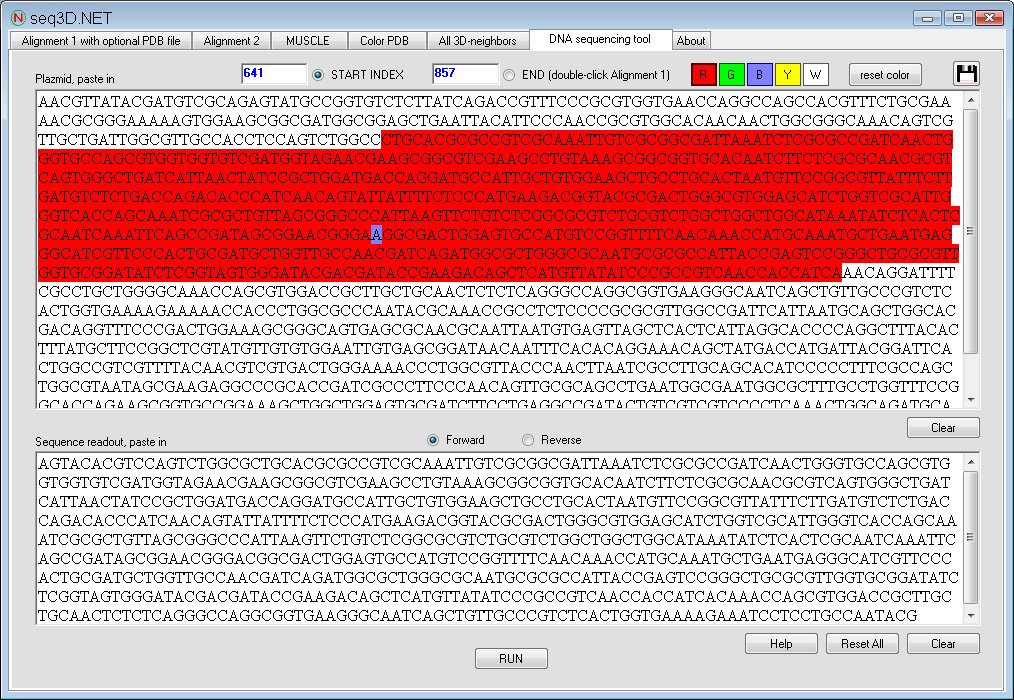
The user provides the plasmid in plain text and the sequencing data, also in plain text. RUN command will create sequence alignment and direct the user to the Alignment 1 window. There, two residues representing Start and End of a first region are double-clicked. On return to "DNA sequencing" tab, Start and End indexes will be automatically filled, and the selected region can by highlighted with one of the four colors. The actions are repeated until the end of a trusted alignment. Sequencing readouts with additional primers (if available) are analyzed and the resulting colored plasmid is saved as an *.rtf file.
This simulated two-primer sequencing result was assembled in under 1 minute from three raw text files (plasmid, sequencing result with primer 1 and sequencing result with primer 2), and shows the presence of a single mutation:
aacgttatacgatgtcgcagagtatgccggtgtctctta
tcagaccgtttcccgcgtggtgaaccaggccagccac
gtttctgcgaaaacgcgggaaaaagtggaagcggcgat
ggcggagctgaattacattcccaaccgcgtggcacaac
aactggcgggcaaacagtcgttgctgattggcgttgcc
acctccagtctggccctgcacgcgccgtcgcaaattgt
cgcggcgattaaatctcgcgccgatcaactgggtgcca
gcgtggtggtgtcgatggtagaacgaagcggcgtcgaa
gcctgtaaagcggcggtgcacaatcttctcgcgcaacg
cgtcagtgggctgatcattaactatccgctggatgacca
ggatgccattgctgtggaagctgcctgcactaatgttcc
ggcgttatttcttgatgtctctgaccagacacccatcaac
agtattattttctcccatgaagacggtacgcgactgggc
gtggagcatctggtcgcattgggtcaccagcaaatcgc
gctgttagcgggcccattaagttctgtctcggcgcgtc
tgcgtctggctggctggcataaatatctcactcgcaatc
aaattcagccgatagcggaacgggaaggcgactggagt
gccatgtccggttttcaacaaaccatgcaaatgctgaat
gagggcatcgttcccactgcgatgctggttgccaacga
tcagatggcgctgggcgcaatgcgcgccattaccgagt
ccgggctgcgcgttggtgcggatatctcggtagtggga
tacgacgataccgaagacagctcatgttatatcccgccg
tcaaccaccatcaaacaggattttcgcctgctggggcaa
accagcgtggaccgcttgctgcaactctctcagggcca
ggcggtgaagggcaatcagctgttgcccgtctcactgg
tgaaaagaaaaaccaccctggcgcccaatacgcaaaccg
cctctccccgcgcgttggccgattcattaatgcagctgg
cacgacaggtttcccgactggaaagcgggcagtgagcg
caacgcaattaatgtgagttagctcactcattaggcaccc
caggctttacactttatgcttccggctcgtatgttgtgtg
gaattgtgagcggataacaatttcacacaggaaacagcta
tgaccatgattacggattcactggccgtcgttttacaacg
tcgtgactgggaaaaccctggcgttacccaacttaatcg
cctt