
SEQMOL index

Multiple sequence alignment
Download
MUSCLE can generate multiple sequence alignments from FASTA formatted protein, RNA or DNA sequences.
Correct FASTA format:
>NAME1
PGHSTERTWYEFDH...
>NAME2
PGHSTEKSRYEFDH...
...
Available MUSCLE settings
1. "max iterations"
This setting may affect the alignment results and should be read about in MUSCLE documentation.
Other settings WILL NOT affect the alignment results, but they will determine how the resulting alignment is analyzed by the program.
2. "do Alignment 2" will feed the results of MUSCLE run to the Alignment 2 tab.
3. "Use S.W. ..." weights sequences to reduce contribution of redundant sequences to the conservation score.
S.W. is always "ON", but when this option is checked, weights for entire sequence are determined by subsequence selected in the Color PDB tab.
4. "Conservation score for sequence number No" sets the FASTA sequence for which the conservation scores will be computed. The output conservation scores,
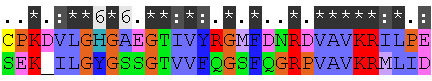
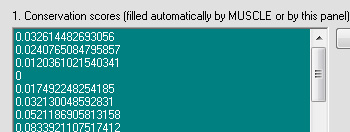
are only correct for selected sequence No. Because sequence No. in the alignment is unknown before the MUSCLE run, the No. can be found from a preliminary MUSCLE run, and then the alignment should be simply re-run with the correct No. entered.
When conservation scores are not needed, settings 2, 3 and 4 can be simply ignored.