SEQMOL index

Linking sequence alignment and PDB coordinates
Download
To enable many useful features of the application, including PDB structure analysis, coloring of PDB by conservation and covariation, exploring sequence alignments + PDB in parallel (including establishing common residue numbering),
PDB file must be provided along with sequence alignment to the "Alignment 1" window by File-Open or drag-dropping,![]() will become
will become ![]()
The sequence of the PDB file's relevant chain should be among the aligned sequences.
PDB file should be in standard format (as output by PyMol, for example),
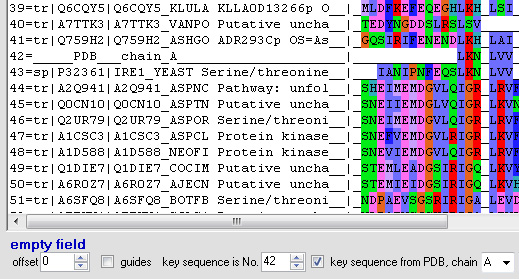
- "key sequence from PDB" should be selected
- number of the PDB sequence in the sequence alignment is the No. of the key sequence
- PDB chain is what was used in the sequence alignment (in this example, PDB chain A is sequence No. 42).
When settings are correct, double clicking a sequence position should return the number of corresponding amino acid in the PDB file: